DNA Methylation Defined
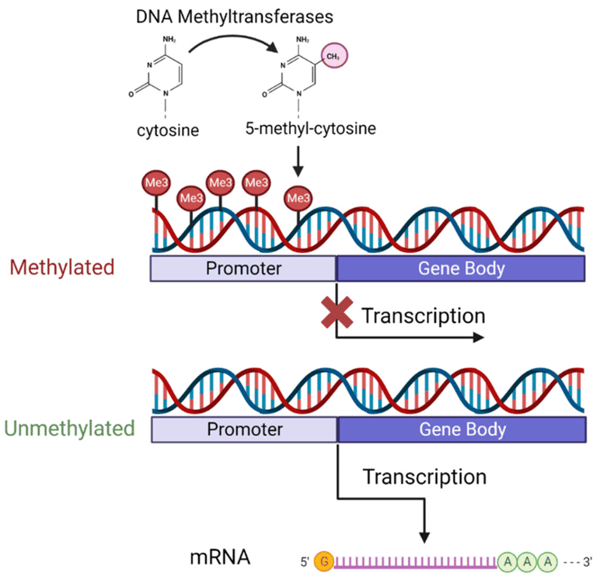
5-methylcytosine (5mC) is a reversible epigenetic mark. In humans and other vertebrates, 5mC is located across the genome and is found primarily in the symmetrical cytosine-guanine (CpG) dinucleotide context.1 5mC (DNA methylation) consists of a methyl group being added to the fifth position of the cytosine ring of DNA. A methyl group is transferred from S-adenosyl-L-methionine to the fifth position of the cytosine ring.2 De novo and maintenance DNA methyltransferases (DNMTs) are responsible for stablishing and maintaining DNA methylation. DNMT1 is responsible for maintenance DNA methylation. Maintenance DNA methylation involves adding a methyl group to hemimethylated strands of DNA.3 DNMT1 localizes to the replication forks during S phase of the cell cycle after recognizing the cofactor ubiquitin-like PHD and ring finger domains1 (UHRF1). The methylation state is then copied by DNMT1 to newly synthesized strands of DNA.4 DNMT3 family members perform de novo DNA methylation. De novo DNA methylation is responsible for establishing DNA methylation and adding a methyl group to unmethylated strands of DNA.5
Source: Biomedicines 2023, 11(7), 1987
In the mammalian genome, 70-80% of the expected 20-25% of CpGs are methylated.6 Most of the CpGs in the mammalian genome are found in intergenic and intronic regions of DNA, which mainly reside in repeat sequences and transposable elements. Transposable elements and pericentromeric satellite repeats are suppressed by DNA methylation. Approximately 90% of DNA methylation is found at repetitive elements.7 Transposable elements are able to invade new regions of the genome. To prevent this, transposable elements are heavily methylated. Improper silencing of transposable elements can lead to increased genome instability and cause chromosomal rearrangements and gene disruption. The significance of DNA methylation at pericentromeric repeats is unclear.
CpG islands are small CG dense regions of about one kilobase. CpG islands are found in the promoters and first exons of genes and are unmethylated. CpG islands are located in open chromatin and are enriched in acetylated histones H3 and H4. Those epigenetic marks represent activation of transcription. However, in unhealthy cells CpG islands are hypermethylated resulting in transcriptional silencing.
Changes in DNA methylation have been seen in many cancers including breast, liver, bone, and colon8. The onset of malignancies has been associated with aberrant changes in DNA methylation. Some changes in DNA methylation associated with malignancies include tumor suppressor genes becoming hypermethylated and oncogenes and repetitive elements becoming hypomethylated. Hypermethylation prevents transcription of tumor suppressor genes while hypomethylation at repeat elements is seen in a variety of cancers and can lead to genomic instability and promote chromosomal rearrangements.
DNA methylation is an important factor in the onset of cancer. Thus, assessment of DNA methylation could help with treatment and prognosis of patients. To assist with your first steps into leveraging methylation in your research, LGC Clinical Diagnostics have developed the first of its kind Seraseq® Methylated ctDNA Mutation Mix and Seraseq® Unmethylated ctDNA Mutation Mix for global analysis of CpG methylation status of ctDNA. To learn more, click here, or on the button below.
Questions?
Email CDx-Sales@lgcgroup.com, call +1.508.244.6400 or 800.676.1881.
Note: This blog article was co-authored by Andrew Anfora of LGC Clinical Diagnostics.
References
- Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011 Jun;2(6):607-17. doi: 10.1177/1947601910393957. PMID: 21941617; PMCID: PMC3174260.
- Menezo Y, Clement P, Clement A, Elder K. Methylation: An Ineluctable Biochemical and Physiological Process Essential to the Transmission of Life. Int J Mol Sci. 2020 Dec 7;21(23):9311. doi: 10.3390/ijms21239311. PMID: 33297303; PMCID: PMC7730869.
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992 Jun 12;69(6):915-26. doi: 10.1016/0092-8674(92)90611-f. PMID: 1606615.
- Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007 Sep 21;317(5845):1760-4. doi: 10.1126/science.1147939. Epub 2007 Aug 2. PMID: 17673620.
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999 Oct 29;99(3):247-57. doi: 10.1016/s0092-8674(00)81656-6. PMID: 10555141.
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982 Apr 24;10(8):2709-21. doi: 10.1093/nar/10.8.2709. PMID: 7079182; PMCID: PMC320645.
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011 Feb;12(2):123-35. doi: 10.1038/nrg2932. Epub 2011 Jan 11. PMID: 21221116.
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999 Feb;21(2):163-7. doi: 10.1038/5947. PMID: 9988266.



