How to get the most from your Seraseq® FFPE Reference Materials
Most tumor profiling assay workflows use tissue biopsy samples that are formalin fixed and paraffin embedded (FFPE), a process that introduces chemical damage in the nucleic acids in the tissue specimen. Depurination, depyrimidination, deamination, oxidation, nicks, and double strand breaks leading to nucleic acid fragmentation may be all found in DNA extracted from FFPE tissue. Cytosine deamination can lead to incorrect sequencing results and may affect accurate identification of lower frequency variants in FFPE DNA samples, but all types of damage affect the sequencing quality (e.g. uniformity of coverage) and downstream genomic analysis.1
In order to assess the effects of nucleic acid damage and extraction method on NGS results and validate the suitability of the analytical workflow for FFPE samples, a full-process FFPE reference material should be included. However, it should be representative of the real-life samples to be useful.1 LGC Clinical Diagnostics developed a broad range of such reference materials for analysis of DNA variants, RNA fusions and complex genomic signatures (TMB, MSI and HRD), including compromised FFPE DNA products that mimic the damage and size profiles of archived tissue biopsy specimens (Figure 2).
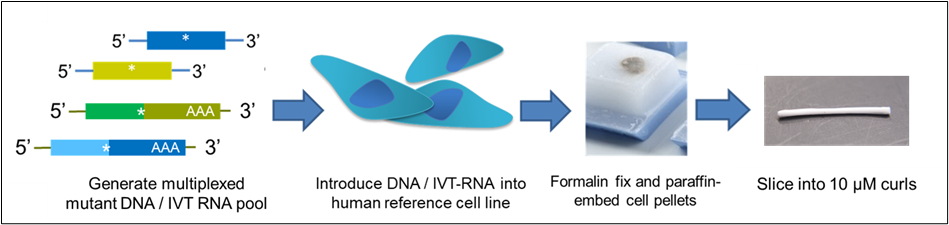
Figure 1. The Seraseq® FFPE reference material design process.
When using both a positive FFPE reference material containing the genomic alterations of interest and a WT negative reference in an experiment, it is important to match the fixation level of the samples, to ensure the results are directly comparable and account for background noise introduced by FFPE-induced DNA damage.
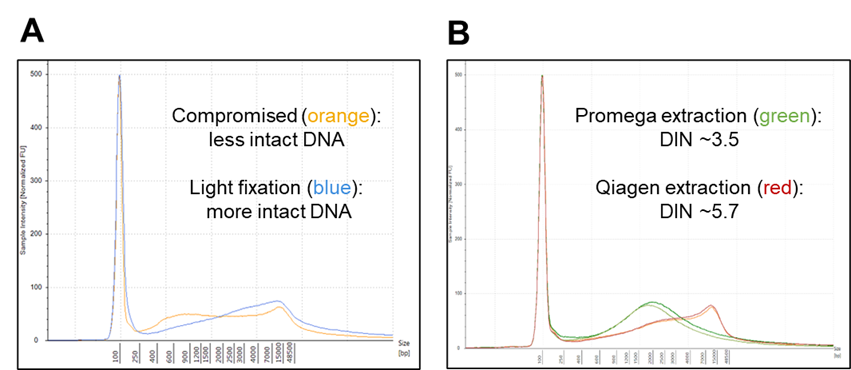
Figure 2. A. Representative DNA size profiles of the two fixation levels: compromised (orange) and standard lightly fixed (blue) Seraseq FFPE products. B. Representative DNA size profiles and DIN values of the Compromised FFPE DNA Tumor Reference material after extraction with the Promega Maxwell® RSC DNA FFPE kit (green) and Qiagen QIAamp® DNA FFPE Tissue kit (red). Average DIN values obtained across all released lots were 3.1 ± 0.8 and 4.8 ± 0.5, respectively. DIN values were measured with the gDNA ScreenTape assay. RNA quality was low for both fixation methods, with measured RIN scores ~1 and DV200 >70% (compromised) and RIN scores ~2 (light fixation) after extraction with the Agencourt Formapure Kit and measured with the Agilent Bioanalyzer RNA 6000 Nano Kit (not shown).
The DNA yield, integrity, sequencing metrics and measured variant allele frequencies (VAFs) obtained from FFPE samples can vary widely depending on the extraction kit used.2 This variation can be due to differences in relative extraction efficiency of the DNA of different fragment lengths, as well as bias in the reversal of formaldehyde damage during DNA isolation with the respective kits.
While the Seraseq® FFPE reference materials have a guaranteed yield of >100 ng DNA or >400 ng RNA per curl, this is only applicable when using the same extraction kit as used by LGC Clinical Diagnostics for product release testing as yield can vary (see Table 1). Although lower yield does not affect the downstream results, maximizing the performance of nucleic acid extraction ensures sufficient sample input into different sequencing protocols. Below are some recommendations that can help prevent low yield and sample loss, with a focus on DNA extraction.
|
FFPE DNA extraction yields (ng) |
|||
|
Material |
QIAamp DNA FFPE Tissue |
Maxwell RSC DNA FFPE |
AllPrep FFPE |
|
Seraseq® Compromised FFPE Tumor DNA |
177 ± 50 375 ± 148# |
384 ± 51 290 ± 82# |
551 ± 151# |
|
Seraseq® FFPE HRD High-Positive |
165 ± 41 |
136 ± 55 |
161 ± 22* |
|
Seraseq® FFPE HRD Low-Positive |
235 ± 68 |
122 ± 39 |
293 ± 44* |
|
Seraseq® FFPE HRD Negative |
505 ± 191 |
200 ± 77 |
315 ± 10* |
|
Seraseq® Lymphoma FFPE DNA |
193 ± 44# |
124 ±11# |
N/A |
|
FFPE RNA extraction yields (ng) |
|||
|
Material |
AllPrep DNA/RNA FFPE |
Maxwell RSC RNA FFPE |
Agencourt Formapure |
|
Seraseq® FFPE Fusion RNA v4 RM |
1021 ± 240 774 ± 178* |
170 ± 28 238 ± 77* |
1177 ± 255 |
|
Seraseq® FFPE WT |
N/A |
N/A |
449 ± 35 |
|
Seraseq® Compromised FFPE WT |
N/A |
N/A |
1278 ± 187 |
Table 1. Variability of DNA and RNA yields obtained for different Seraseq® FFPE Reference Materials. For all values, ± is one standard deviation; Unless otherwise indicated, data is representative of a single lot of material. #average internal data from all released lots *data from an external lab.
Steps for Success in Extracting DNA from Biosynthetic FFPE Samples
The Seraseq® sample pellets are smaller and more transparent than those from clinical samples and can be difficult to see. It might help to take the following precautions:
- Extract the Seraseq® sample alongside a clinical one to track the sample visually.
- Mark the outside of the tube and the cap and place in a centrifuge so that the mark is facing the outside of the rotor (where the pellet is expected to be).
- When using a protocol requiring discarding a supernatant, save the supernatants during the first few runs if in any doubt as to the location of the pellet.
DNA yield extracted using automated protocols tends to be lower than with corresponding manual protocols. When using an automated extraction system, we always recommend validating new automated instrumentation with reference materials such as Seraseq® WT FFPE materials.
QIAamp® DNA FFPE Tissue Kit (Qiagen)
- 1 mL xylene can be added directly to the product tube vial containing the curl (Figure 3A) to avoid transferring the curl. It might be helpful to remove the product tube label to better see the contents of the tube If transferring the curl to a new vial is necessary, we recommend trying to tap it over with the product vial on top of the other vial and avoid using forceps. Alternatively, you may be able to solubilize the curl first in the original tube and transfer the entire contents to a new tube.
- When removing xylene after deparaffinization, be conservative. Remove 900 uL only and leave the rest in the vial to avoid disturbing the pellet. Air dry in a chemical hood before adding the ethanol.
- The DNA will only become visible after adding the ethanol to precipitate. If it’s not readily visible, invert the vial a few times after vortexing to visualize it. The precipitate should appear as a white, gelatinous mass (Figure 3B). After centrifugation, the precipitate should be readily visible as a pellet at the bottom of the tube (Figure 3C).
- After centrifugation of the precipitate, remove as much of the supernatant as possible, but be careful to avoid disturbing and / or aspirating the pellet. Using a smaller size pipette could be helpful.
Maxwell® RSC DNA FFPE Kit (Promega)
- Melting and deparaffinization incubation temperatures are critical; too high a temperature will result in reduced DNA yield due to excess single-stranded DNA in the sample. Sealing the vial tightly is also important.
- There is no pellet so the risk of losing the sample is lower. However, care must be taken when removing the aqueous layer after the RNAse A treatment, to avoid transferring any of the denatured protein interphase, as it can clog the cartridge and reduce yield.
AllPrep® DNA/RNA FFPE Kit (Qiagen)
- Follow similar overall precautions as with the QIAamp® DNA FFPE Tissue kit
- When resuspending the pellet in buffer PKD before proteinase K treatment, it can be visualized by adding the buffer forcefully to dislodge it.
- If not extracting RNA at the same time, be conservative when removing the supernatant to avoid losing DNA.
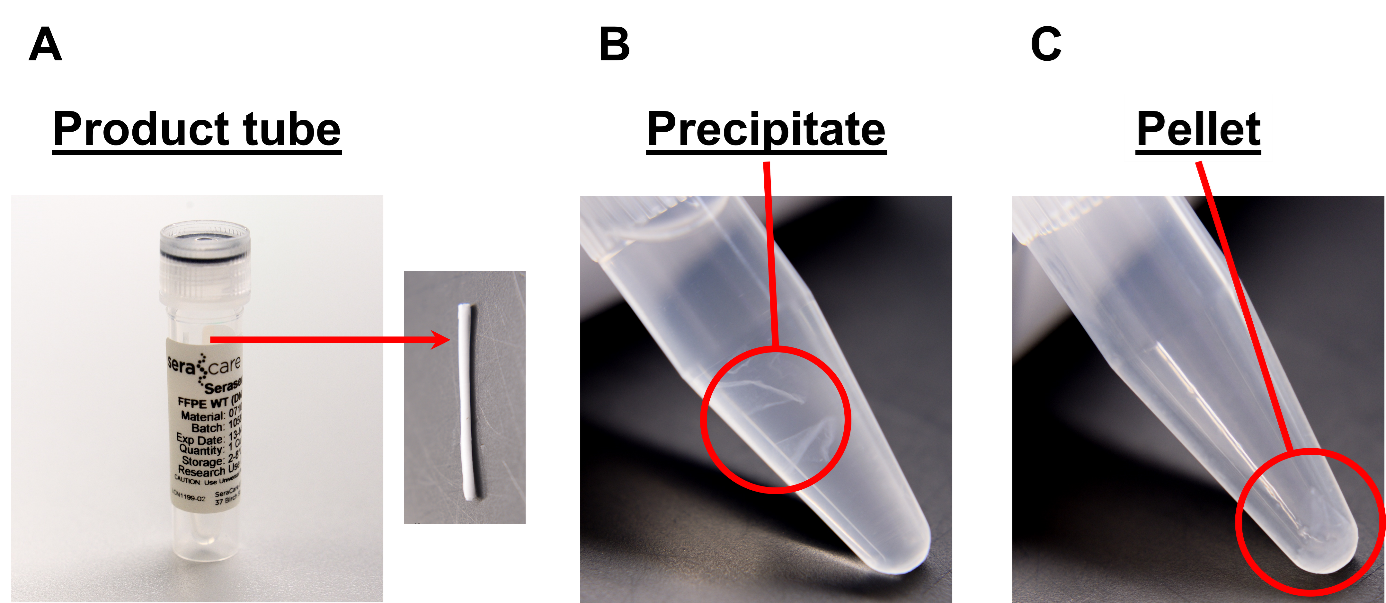
Figure 3 (Ed Davis). Seraseq® FFPE material at different stages of extraction. A., Left: Original product tube, within which the curl is contained. Right: Curl after removal from the tube. Typical dimensions are 35mm x 25mm x 10μm. B. Nucleic acid precipitate following ethanol addition. C. Pellet at bottom of tube following centrifugation of the precipitate.
Learn More
Check out our portfolio of Seraseq® FFPE Reference Materials by clicking here, or on the button below.
References:
- Steiert, T. A., Parra, G., Gut, M., et al. (2023). A critical spotlight on the paradigms of FFPE-DNA sequencing. Nucleic Acids Research, 51(14), 7143–7162. https://doi.org/10.1093/nar/gkad519
- Bonnet E, Moutet M-L, Baulard C, et al. Performance comparison of three DNA extraction kits on human whole-exome data from formalin-fixed paraffin-embedded normal and tumor samples. PLoS One. 2018; 13 (4); e0195471.